Google launches 'AlphaGenome,' an AI that can analyze the effects of genetic mutations, capable of inputting 1 million base pairs at once and potentially helping to establish treatments for genetic diseases
The DNA of all living organisms, including humans, contains hundreds of millions of four types of nucleotides: adenine (A), thymine (T), guanine (G), and cytosine (C). The arrangement of these nucleotides determines the diverse characteristics of the organism. This ATGC arrangement is called the 'base sequence,' and if part of the base sequence mutates due to some factor, it can cause genetic diseases and other problems. A research team at Google has developed an AI called ' AlphaGenome ' that can analyze these 'effects of changes in base sequences.'
AlphaGenome: AI for better understanding the genome - Google DeepMind
In September 2023, Google announced ' AlphaMissense ,' an AI that predicts the harmfulness of genetic mutations. While AlphaMissense targeted the 'regions of the genome that describe what proteins are produced,' the newly announced AlphaGenome is unique in that it can also target 'non-coding regions' that do not function as protein blueprints. Non-coding regions make up a large portion of the genome and were not previously considered very important, but recent research has revealed that they have important functions, such as 'regulating the on/off of genes.'
AlphaGenome is built using ' Enformer ', an AI architecture for genetic research developed based on ' Transformer '. It also uses genetic information databases such as ' ENCODE ', ' GTEx ', ' 4D Nucleome ', and ' FANTOM5 ' for training data.
AlphaGenome can input up to 1 million base sequences at once, enabling comprehensive analysis of complex gene regulation across multiple steps. It is also the first AI model to support analysis of the effect of gene mutations on RNA splicing .
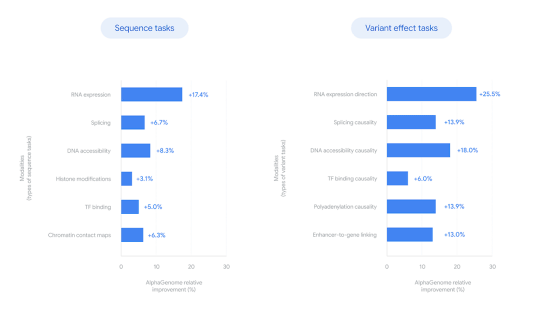
Below is a graph comparing the benchmark results of AlphaGenome and existing genetic analysis AI models. AlphaGenome shows significantly higher performance than existing models.
Google provides the AlphaGenome API to non-profit researchers, and claims that it can be used to identify the causes of genetic diseases and develop the field of DNA synthesis. However, there are problems with the current AlphaGenome, such as 'it becomes difficult to predict the effects when it is about 100,000 base pairs away from the regulatory factor' and 'the results at the molecular level can be predicted, but the overall picture of how it is related to traits and diseases cannot be predicted.' The research team is working to solve these problems.
Below is the API introduction page for AlphaGenome. The introduction page includes links to various documents and contact information for Google.
AlphaGenome
https://deepmind.google.com/science/alphagenome/
Code and documentation related to AlphaGenome are available at the following links:
GitHub - google-deepmind/alphagenome: This API provides programmatic access to the AlphaGenome model developed by Google DeepMind.
https://github.com/google-deepmind/alphagenome?tab=readme-ov-file
Google has also set up a community forum for researchers using AlphaGenome.
AlphaGenome
https://www.alphagenomecommunity.com/
In addition, a pre-peer reviewed paper describing the AlphaGenome is available at the following link:
AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model
(PDF file) https://storage.googleapis.com/deepmind-media/papers/alphagenome.pdf